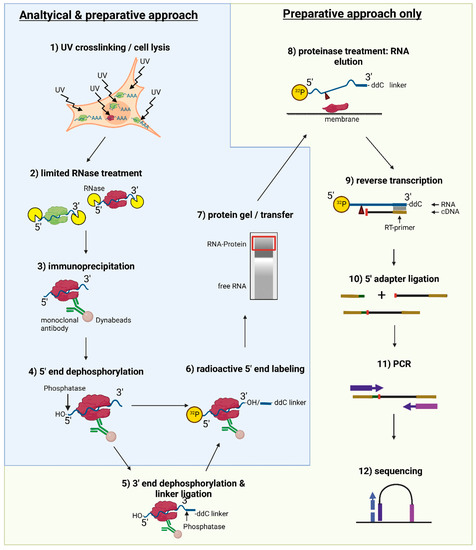
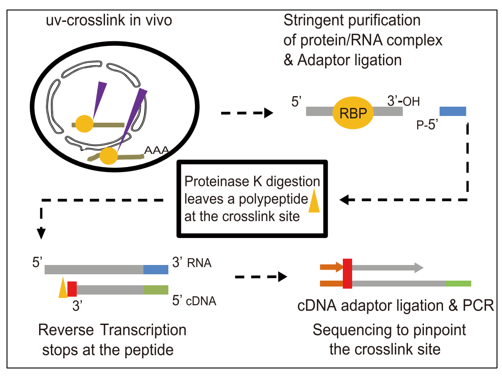
tRIP‐seq reveals repression of premature polyadenylation by co‐transcriptional FUS‐U1 snRNP assembly | EMBO reports

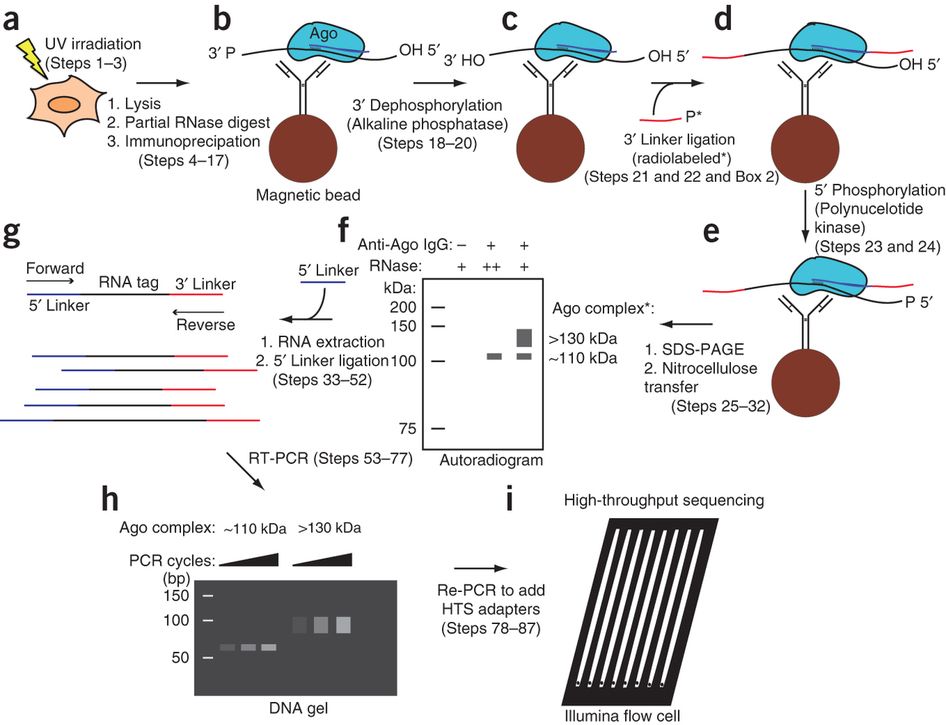
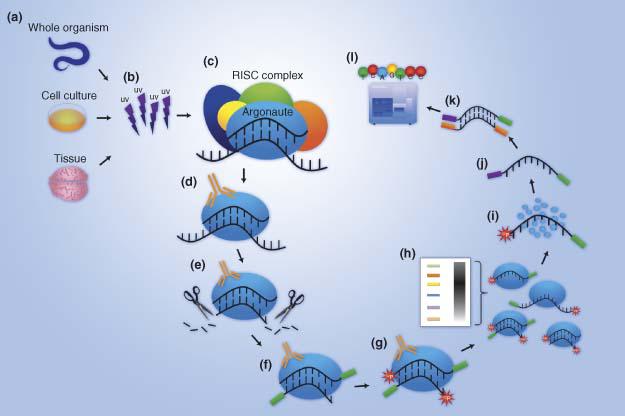
ACE-score-based Analysis of Temporal miRNA Targetomes During Human Cytomegalovirus Infection Using AGO-CLIP-seq
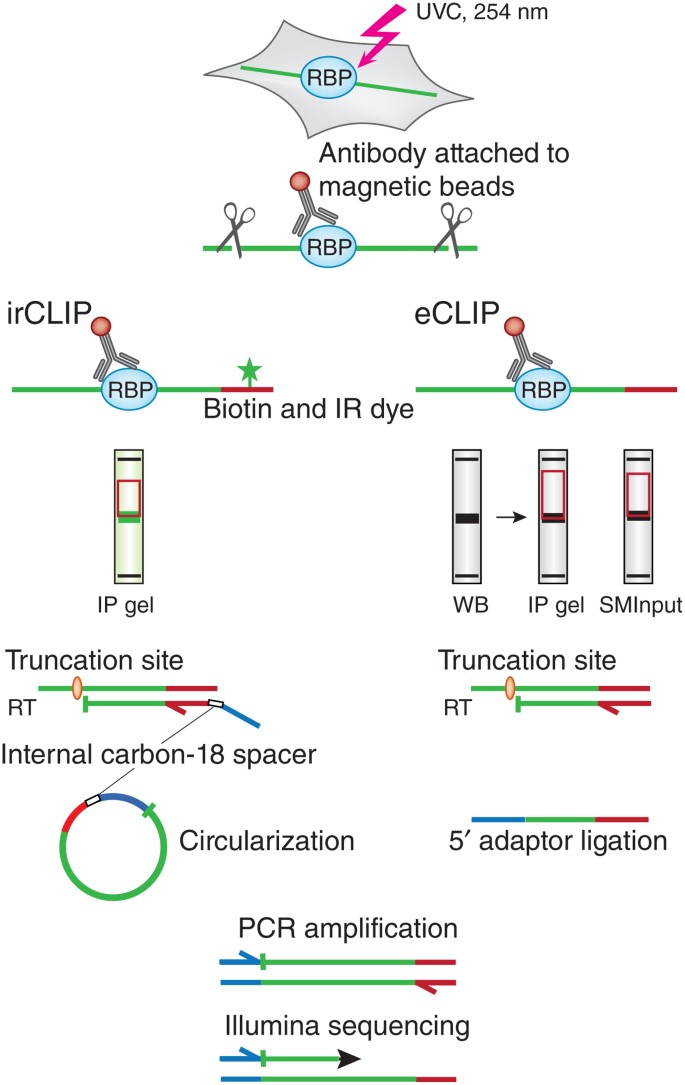
Enhanced CLIP Uncovers IMP Protein-RNA Targets in Human Pluripotent Stem Cells Important for Cell Adhesion and Survival
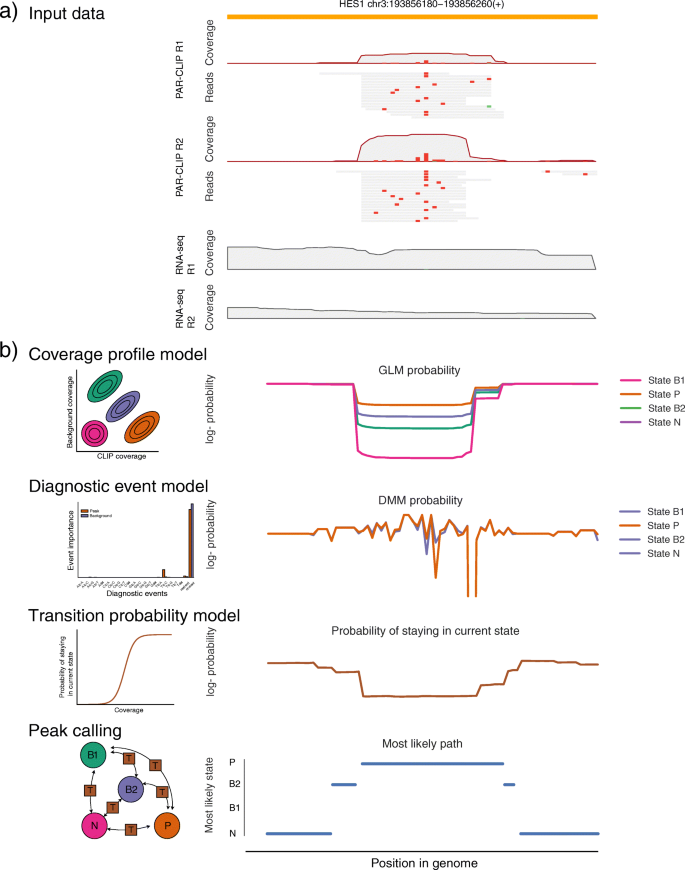
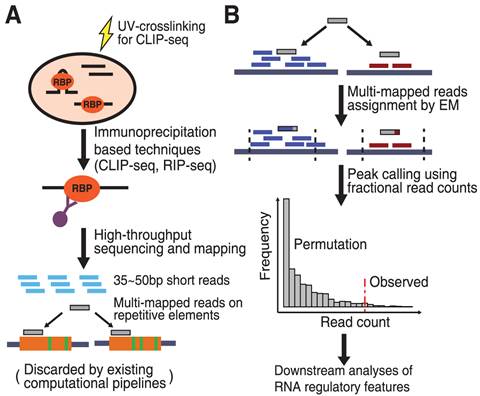
omniCLIP: probabilistic identification of protein-RNA interactions from CLIP -seq data | Genome Biology | Full Text

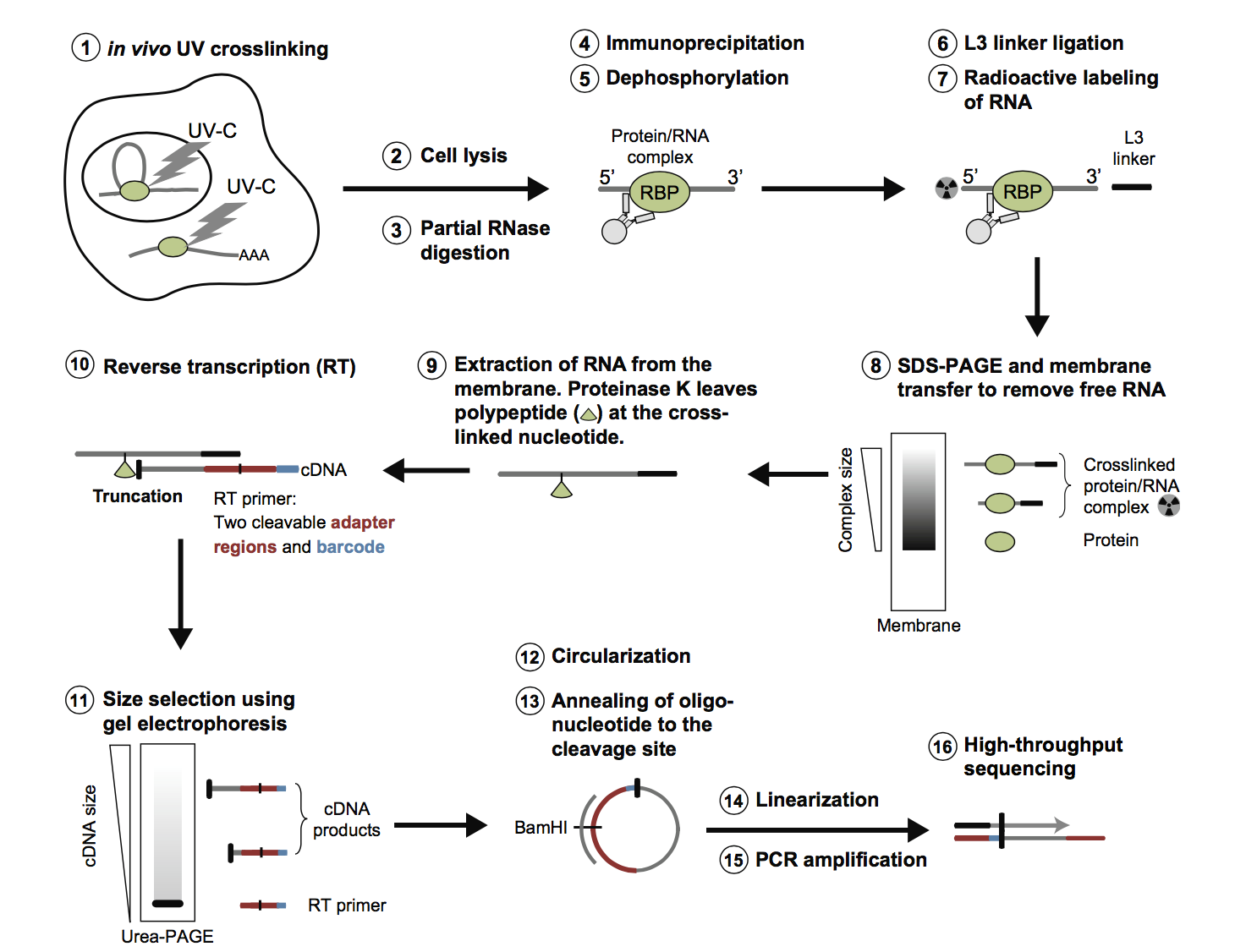
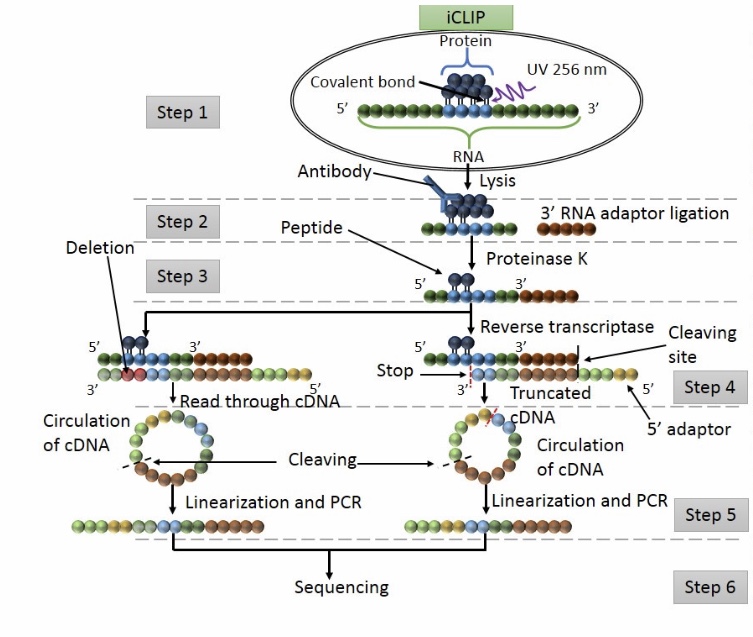
Schematic summary of CLIP and iCLIP methods. After UV cross-linking... | Download Scientific Diagram

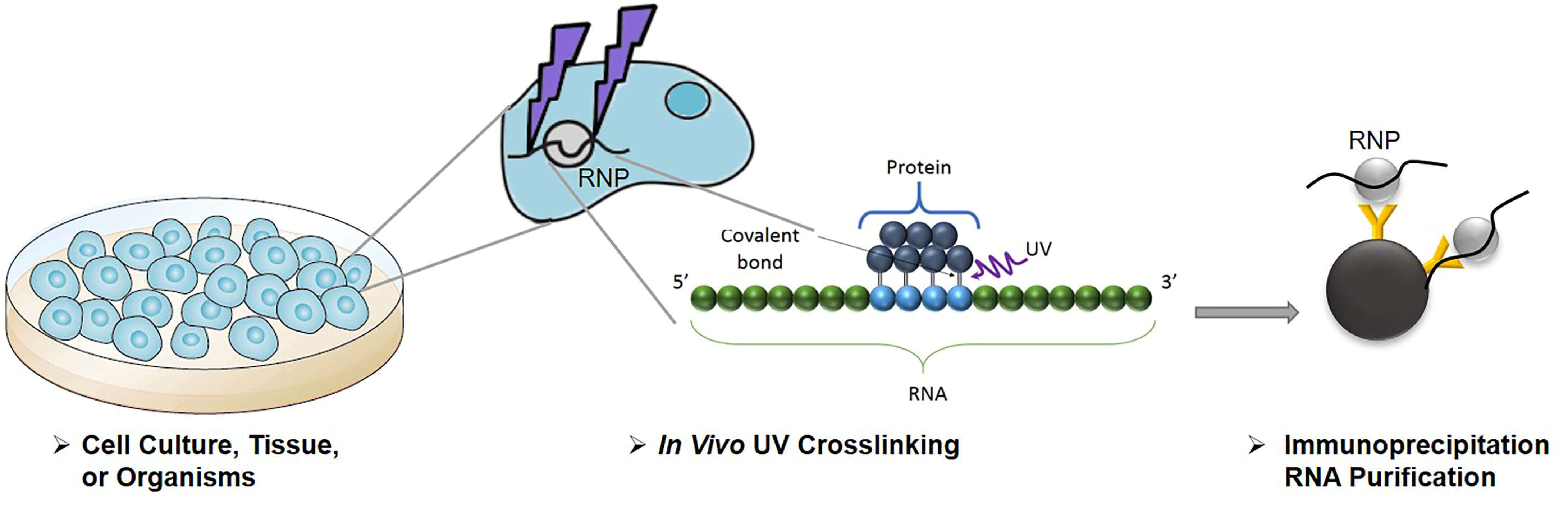
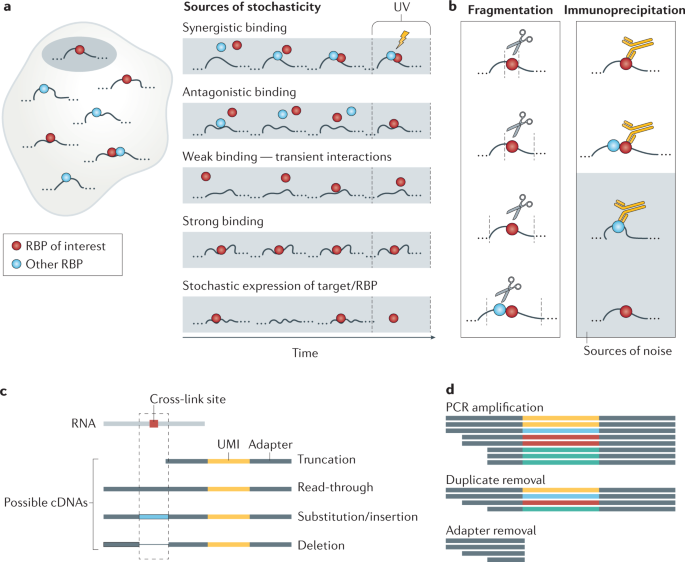
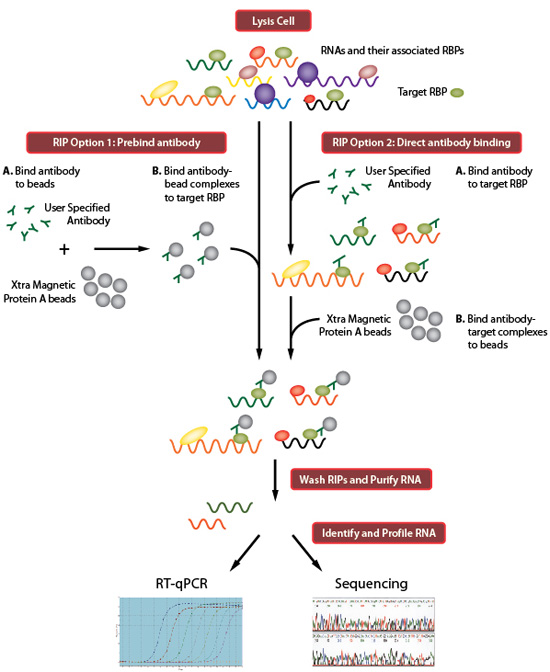
Computational approaches for the analysis of RNA–protein interactions: A primer for biologists - Journal of Biological Chemistry

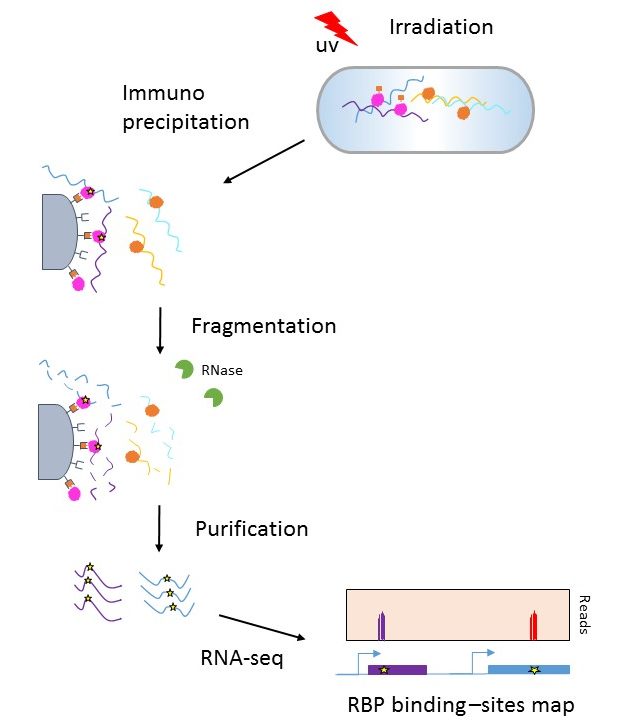
Global identification of RsmA/N binding sites in Pseudomonas aeruginosa by in vivo UV CLIP-seq | bioRxiv